
UNDERSTANDING IMMUNE RESPONSES TO SARS-COV-2
Charu Kaushic | July 6, 2020
Charu Kaushic M.Sc. PhD is the Scientific Director, CIHR-Institute of Infection and Immunity and a Professor in the Dept. Pathology and Molecular Medicine, McMaster Immunology Research Center (MIRC).
Summary
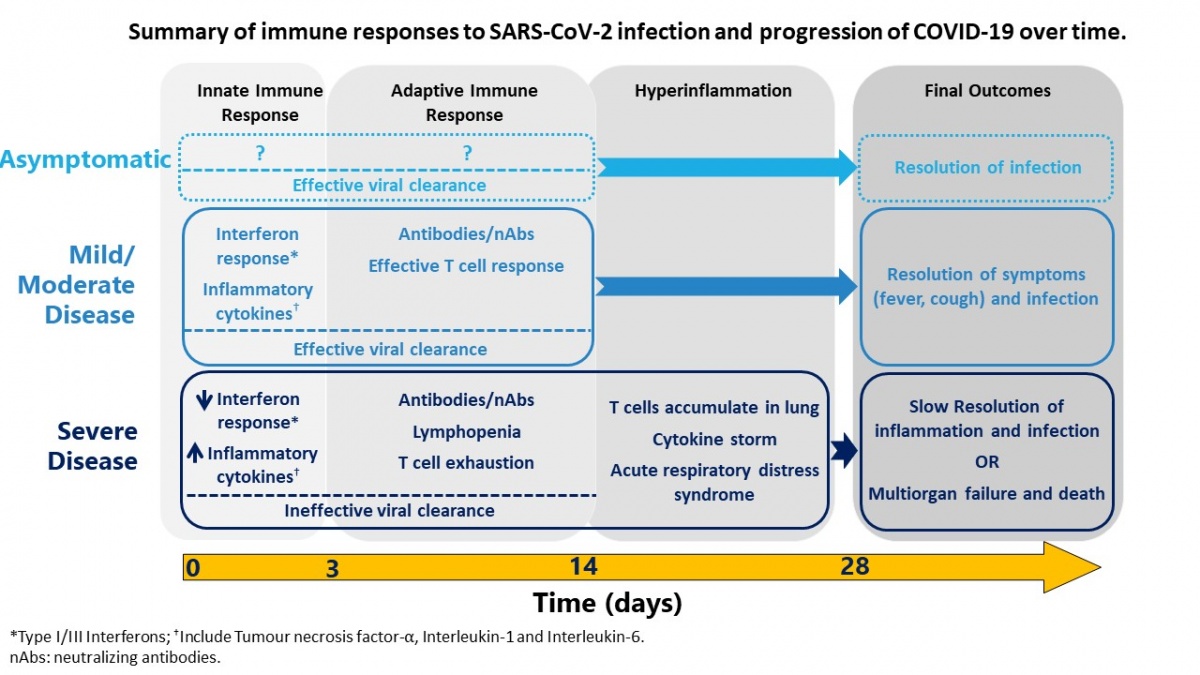
SARS-CoV-2 is a newly emergent coronavirus that is highly infectious and is the cause of COVID-19. The disease caused by the virus is characterized primarily by mild to severe respiratory symptoms that can lead to hospitalization and death in ~1% of cases. Diagnostic tests for detection of the virus in the airways were developed quickly, within weeks of the outbreak, and have played a key role in public health strategy used globally to test, track and isolate cases, likely saving millions of lives worldwide. However, a lot more research is needed to understand this new virus, the disease, and the immune response that could potentially protect, or contribute, to COVID-19. Based on rapidly emerging data, SARS-CoV-2 infection induces differential disease, which is likely the outcome of the interaction between the virus and host immune response (Figure). The most severe disease outcomes are correlated with an over-exuberant immune response. The role of antibodies in immune protection is still not completely understood, making predictions around herd immunity uncertain and utility of “immunity passports” difficult to assess. The final exit strategy from the current pandemic will very likely involve a combination of effective therapeutics to decrease the burden of disease and vaccines to prevent transmission.
1. Immune responses to viruses: innate versus adaptive immunity
Viruses are submicroscopic parasites that require host cells to replicate. These tiny organisms are composed of basic genetic information in the form of nucleic acids (RNA or DNA) packaged within a shell made of protein, called the capsid. Many viruses also have an outer shell called envelope, a lipid membrane containing viral glycoproteins, that facilitates the entry of the virus into host cells. The main function of this whole viral structure is to infect cells, take over the host cellular machinery to multiply and produce lots of virions, that are subsequently released and repeat the process in other cells. While viruses can infect a wide range of animals and plants, most have a narrow range of hosts, and a few can infect different species. Coronaviruses are among the latter category1. Upon infection, the host detects the virus as a “foreign’ entity and mounts a defense through its immune system2. The human immune system is a complex network of cells and organs that can detect and defend against harmful organisms through a variety of mechanisms.
The human immune response to a virus can be divided into different phases, starting within minutes of a viral infection, and lasting over weeks, months, or sometimes years3. The acute phase of the immune response lasts ~72 hours and is dominated by “innate immunity”, which consists of cells and processes that can be deployed quickly and act rapidly to contain the virus and stop its replication within the host. Every cell in the human body is equipped to recognize and react to viruses, this is especially true for the cells that line the mucosal surfaces, the moist inner lining of the body that covers the airways, gastrointestinal and urogenital tracts and other organs. When a respiratory virus, such as coronavirus, encounters the epithelial cells that line the airways, it attaches to specific receptors present on the host cell surface with the help of the viral envelope glycoproteins. This facilitates the entry of the virus into the cell, where it can start replicating by hijacking the cellular machinery. At the same time, the host cell has a variety of receptors on its cell surface and in the cytoplasm, called pattern recognition receptors [PRRs], that recognize the virus and initiate innate immune responses. These include the initiation of Type I and III Interferons (IFNs) and cellular factors that have anti-viral effects, as well as inflammatory factors that send alarm signals and bring in immune cells such as macrophages, dendritic cells and natural killer (NK) cells to the area of infection. The anti-viral and inflammatory milieu as well as immune cells that comprise the innate immune response primarily aim to a) destroy the virus or limit its replication, and b) activate the next phase of the immune response within 48-72 hours. The second phase of immune response is adaptive or acquired immunity which is maintained from weeks to years and involves special organs and tissues like lymph nodes and specialized immune cells such as B and T cells. The adaptive immune response is slower but can customize the immune responses with high specificity to a large variety of pathogens, such as harmful bacteria and viruses. The adaptive immune response is capable of targeting novel viruses that the host has never encountered, and it is also responsible for the “memory” of the immune system, allowing hosts to respond much more rapidly to re-infections even years later.
Vaccines take advantage of this ability to customize a specific memory response, using weakened viruses or viral parts, such that when the host encounters a fully infectious virus, a rapid secondary immune response is mounted that helps resist the infection. The adaptive immune responses have two main components: 1) cellular immunity comprised of T lymphocytes that can attack and kill the infected cells directly or indirectly and 2) humoral immunity which is mediated by B lymphocytes and antibodies, which are complex proteins secreted by plasma cells (differentiated B lymphocytes). Antibodies are found in blood and body fluids, and perform a range of functions such as inhibiting the virus binding, neutralizing the virus, or activating immune cells to kill the virus or infected cells. The characteristic feature of antibodies lies in the ability to recognize and bind to very specific epitopes of the virus or “antigens”. Although antibodies may only last in blood stream for a few days to a few weeks, the plasma cells can keep replenishing them for months. Antibodies belong to five different classes, IgD, IgM, IgA, IgE and IgG, based on the type of protein chains they are made of. They all have slightly different structures, characteristics, and functions. IgM, IgG and IgA antibodies all have anti-viral functions, but IgG antibodies are the longest lasting, are present in high amounts in both blood and body fluids and perform a whole range of anti-viral activity.
In summary, the human immune system responds to viruses in a variety of ways, starting with innate immunity which is composed of rapid and broad responses followed by adaptive immunity that is mediated by specialized immune cells and antibodies and is characterized by specificity, diversity and memory. The outcome of a viral exposure is determined by the sum total of the ability of the virus to subvert and overcome host responses and the ability of the host to mount rapid and potent anti-viral immune responses.
2. Immune responses to SARS CoV-2: current state of knowledge
The coronaviruses belong to a large family of RNA viruses that can infect several mammalian species including cats, bats, camels, cows, pigs and humans4. There are seven coronavirus species capable of infecting humans, all of which cause respiratory infections. Four of these cause colds (alpha CoV, 229E, NL3 and beta CoV, OC43, HK41) that vary in severity but rarely cause fatality. Three coronaviruses that cause much more serious respiratory infections and high mortality rates5,6 have crossed from other species into humans in the last two decades: Severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1), Middle East respiratory syndrome (MERS)-CoV, and SARS-CoV-2. SARS-CoV-2 causes COVID-19 which is causing widespread morbidity in the current pandemic.
The coronaviruses have a relatively simple structure: a single stranded RNA genetic core surrounded by a capsid and an outside envelope. Four structural proteins make up the virus: the nucleocapsid (N), the membrane (M), the envelope (E) and the spike protein (S)7. The S protein forms trimers which project out from the viral envelope like a crown, hence the name “coronavirus”. The spike protein plays a crucial role in infection, since it contains the receptor binding domain (RBD) that facilitates viral attachment to the host cell. The S proteins of both SARS-CoV-1 and SARS-CoV-2, that are closely related to each other, bind to a common protein found on the surface of cells, called Angiotensin converting enzyme 2 (ACE2), to enter host cells8,9. ACE2 is commonly expressed in cardiopulmonary (heart and respiratory) tissues, the gastrointestinal tract, immune cells (monocytes, macrophages and dendritic cells) and other tissues, with the main function of regulating blood pressure10. The S protein is the immunodominant protein, meaning it gives rise to potent T cell and antibody responses and is an attractive target for vaccine and therapeutics because of its involvement in viral attachment and entry. However T and B cells are capable of targeting the M and N proteins as well11.
Although few studies have examined innate immunity to SARS-CoV-2, thus far it appears that like other coronaviruses, it induces a range of innate immune responses, including Type I/III IFNs, the primary anti-viral factors made by cells, and proinflammatory cytokines like TNF-⍺, IL-1 and IL-612. Early studies indicate that SARS-CoV-2 suppresses Type I/III IFNs to a greater extent than SARS-CoV-113. However, it does induce potent proinflammatory cytokine response14 and this imbalance between low innate anti-viral response and high inflammation may contribute to development of the new coronavirus disease 2019 (COVID-19). Indeed, IFN antagonism correlated with increasing severity of COVID-1915.
Information regarding the adaptive immune responses against SARS-CoV-2 is starting to emerge. Although T cell responses against the S and N proteins of the virus have been observed in most clinical studies, patients with more severe disease appear to have significant lymphopenia with drastic decreases in T cell numbers in the blood16,17. T cells in patients with severe COVID-19 are activated but show mainly inflammatory responses as well as characteristics of exhaustion18, whereas individuals with mild disease show more effective, multifunctional T cells, with CD8 T cells able to kill virus-infected cells and CD4 T cells producing a variety of cytokines that help other aspects of immune responses19,20. In general, the T cell responses to the virus are biased towards inflammation which may contribute to the severity of COVID-19. Other studies have shown that in severely sick patients, while T cells are decreased in the blood, large numbers accumulate in the lungs and likely contribute to the acute respiratory distress syndrome (ARDS)21,22 by inducing a cytokine storm characterized by uncontrolled levels of inflammatory cytokines (cytokine release syndrome)23 (discussed in Section 7).
The humoral response to SARS-CoV-2 is being studied extensively, given the expected importance of antibodies in protection from re-infection. Several studies have now shown that SARS-CoV-2 infection leads to a robust antibody response within 7-14 days as IgM, IgG and IgA have been detected in almost all infected individuals and that IgG antibodies persist in the weeks following recovery24. The antibodies most commonly detected are against the SARS-CoV-2 N and S proteins25,26. Antibodies against the RBD of the S protein are of interest because they can prevent the virus from attaching to the entry receptor, ACE2, on host cells. RBD-specific antibodies, which are very effective neutralizing antibodies (nAbs) have been detected in most patients27,28. What is less clear is whether all infected individuals make high amounts of these nAbs, how long the antibody response will persist, and whether the nAbs correlate with protection. Studies of other coronavirus infections showed that less severe infections and mild disease led to short-term antibody responses29,30 whereas SARS-CoV-1 and MERS, which cause more severe disease, led to longer-term responses that lasted for 2-3 years29,31,32, with some reports showing even longer memory33.
3. Testing for immune responses versus testing for virus
Given that SARS-CoV-2 is a newly emerged coronavirus that spread at an unprecedented speed across the world, it has been a race against time to develop diagnostic tests. Within the first few weeks of the infection spreading in Wuhan, it became increasingly clear that controlling the spread of this new virus was going to be very challenging, because it is highly infectious, not everyone who gets infected develops full blown disease, and the virus can be transmitted by people with very mild symptoms or no symptoms. In the last few months, epidemiological data has shown that the average number of people infected by one person, if no control measures are in place (known as R0 [R naught]) for SARS-CoV-2 is around 2.2-2.634–37. This means that the virus is more infectious than flu (R0, 1.838) and less than measles (R0, 1539). Although asymptomatic transmission rate varies significantly depending on geographical locations and the public health measures in place in different countries, some estimates have put the contribution of asymptomatic transmission to overall number of cases as high as 40-45%40.
Molecular Tests: Within weeks of COVID-19 emergence, the first diagnostic tests to be developed were molecular tests based on nucleic acid amplification. These are PCR (polymerase chain reaction)-based tests that rely on detecting viral genetic material isolated from throat or nasal swabs. Sputum or bronchoalveolar lavage (BAL) samples can also be used, if infection has spread to the lungs41. The basic process involves isolating viral RNA from the biologic samples, converting it into DNA using reverse transcriptase enzyme and then amplifying the DNA by PCR, using SARS-CoV-2-specific primers. The methods for these assays were quickly shared and published, and have become widely used around the world42,43. While this type of testing is labour intensive, it can detect active infection with high sensitivity44. In the absence of vaccine and therapies, this test has played a key role in the global public health strategy to keep SARS-CoV-2 infections in check, using rapid testing, tracing and isolation. The limitation of this assay is that it is typically used for diagnosing active infection in symptomatic individuals. Given that the virus can be transmitted asymptomatically, it is challenging to be able to trace every infected person using this test.
Serologic Testing: This type of testing detects the presence of antibodies against SARS-CoV-2 in the blood of individuals45. Antibodies develop almost universally following infection with the virus, in all individuals who are not immunocompromised, within 1-2 weeks after infection24. Therefore, it can not only confirm viral infection in people who developed symptoms, but also those with asymptomatic infection. Because the antibodies are likely present in blood for weeks to months after infection, these tests can be performed long after recovery to identify individuals who have previously been infected with SARS-CoV-2. However, there is a lag between the start of infection and the appearance of antibodies in the blood, therefore serology tests cannot be used reliably diagnose active infection. Many of the commercially approved serology tests are called lateral flow assays (LFA), based on immunochromatography technique, which involves running the fluid containing antibodies (patient blood) over a solid substrate containing SARS-CoV-2 antigens. If the antibodies are present in the blood, they will bind the viral protein and cause a color change indicating a positive test. This type of test, based on simple positive or negative detection of antibodies, is useful for large scale surveillance, but does not provide any information regarding the amount, type, or function of the antibodies. A better test for accurately detecting antibodies against SARS-CoV-2 is the enzyme-linked immunosorbent assay (ELISA), a common laboratory test that can measure not only the presence but also the titer (amount) and type (IgG, IgM, IgA) of antibody. This test allows for a better measure of the strength of the humoral response46. In general, the higher the antibody titer the better the protection. The ELISA assays are more complex and can only be performed in a laboratory setting, while the LFA assays are being promoted as easy to perform, inexpensive, point of care tests. However, many of the LFA assays are currently being examined more carefully to ascertain their sensitivity (how frequently they miss positive samples) and specificity (how often they incorrectly diagnose negative samples as positive), both of which have important public health consequences. If the presence of antibody is indeed confirmed to protect against re-infection, then performing serological testing in large populations (sero-surveillance) can play an important role in determining the level of population-based immunity (herd immunity) and validate the concept of an “immunity passport”47.
Viral neutralization assay: The mere presence of antibodies in blood, measured by LFA or ELISA tests, does not show they can effectively block viral infection. The gold standard of testing the ability of an antibody to block or neutralize the virus is viral plaque reduction neutralization assay48. When viruses replicate inside cells grown in cultures, they are subsequently released when the cells are lysed or killed. This results in holes known as “plaques” that can be easily counted under the microscope or quantified using automated end points. If the antibody blocks the ability of the virus to infect the cells, there will be fewer or no plaques that form after a mixture of antibodies and virus are layered on the cells – indicating that the antibody can “neutralize” the virus. This assay measures not only the titer of the antibody but also its ability to protect against viral infection. However, these assays are very labour intensive and, because they involve the use of live infectious virus, must be performed in biosafety level 3 (BSL3) labs48. Alternate tests that use substitute viruses (pseudovirus neutralization assays) have been developed but are currently only being used in research laboratories.
In summary, a number of tests have been developed quickly and adapted from current tests to provide a variety of ways to test SARS-CoV-2 infection as well as host antibody responses. The diagnostic tests that detect presence of virus are complementary to tests that measure host immunity because together they can diagnose current as well as asymptomatic infections. Although not all tests are completely optimized at this point, they have played a critical role in the current public health strategy of testing, tracing, and isolating which has effectively saved millions of lives across the world.
4. The antibody response to SARS-CoV-2 and population seroprevalence
While most studies examining antibody responses have found that almost all individuals infected by SARS-CoV-2 develop antibodies to the virus, it is still unknown how long these antibody responses last and whether the antibodies protect from re-infection. While some individuals developed an antibody response as early as 8 days after infection, in most studies, it peaked around 14 days, with significant variation among patients24. IgM was the first antibody to be detected within the first week, but its levels decreased after 30 days while IgG levels increased slowly but remained high up to 25 weeks post-infection. The antibody titers correlated with the severity of the infection, with higher antibody levels in individuals who developed more severe symptoms. The majority of the antibodies detected were found to be against the N and S proteins of SARS-CoV-2. In particular, antibodies against the RBD of the S protein, which is highly immunogenic, have the potential to be highly neutralizing27,28. Studies found that while in early phase of the immune response, the antibody titer correlated with neutralization, the neutralizing antibodies declined rapidly following recovery49. Previous studies have shown that coronaviruses such as SARS-CoV-1 and MERS that cause more severe infections lead to longer and more potent antibody responses compared to coronaviruses that cause milder ones29,31,32. Thus far there are few reports of re-infection among individuals recovered from COVID-19, but longitudinal studies need to be done to examine how long the protection will last and whether individuals who had mild symptoms will have shorter-term protection compared to those who developed severe disease.
It has been estimated that 60-70% of the population would need to develop antibody responses before herd immunity could develop against SARS-CoV-250. Sero-epidemiological studies of EU/EEA member states show low levels of seropositivity ranging from ~1% (Finland51, Scotland52) to 5% in Spain53 and 8.5% in UK54. This is consistent with seroprevalence studies done in other countries as well55–57 indicating that overall, less than 10% of the population in most countries has been exposed to the virus. Global efforts are ongoing to determine an accurate seroprevalence of SARS-CoV-2 antibodies. However, current estimates suggest that it is highly unlikely that herd immunity will be reached anytime soon in any country around the world.
5. Antibodies as therapies
Neutralizing antibodies that block viral entry have the potential to be used both for therapy and prophylactic application. Transfer of convalescent plasma, which involves transfusion of the liquid portion of blood from individuals who have recovered from an infection into people who are infected, has been tried as a life-saving therapy for many infectious diseases with varying degrees of success. The plasma of recovered individuals contains a variety of antibodies, including nAbs, against the pathogen, which can block infection of new cells in the recipients, allowing for recovery, although the immunity conferred by this type of treatment is short term. The few studies looking at the effectiveness of convalescent plasma transfusion for individuals with severe COVID-19, show varying results58–62, probably because the success of this approach depends on the presence of high titers of nAbs and the timing of the treatment. There are also additional considerations when using convalescent plansma transfusions, including screening for other infectious agents, matching the blood types and potential concerns about the presence of antibodies that could exacerbate disease due to their ability to induce inflammation via antibody dependent enhancement (ADE). Administration of monoclonal antibodies (mAbs), derived from a single plasma cell which produces one type of antibody and results in greater reproducibility and efficiency in biological activity, is an alternative to the use of convalescent plasma, which overcomes most of these limitations63,64. Significant advances have been made in rapidly identifying and producing mAbs which can neutralize viruses. Newer approaches involve isolation of single antibody producing cells from recovered patients, immortalization of the cell followed by rapid scale up of antibody production and testing. This method was successfully used to develop mAb114, which comprised a single antibody, and RGEN-EB3, which contained a cocktail of three-antibodies, against the Ebola virus. Both treatments were shown to be effective reducing mortality in randomized trials65. There are several reports of successful isolation of nAbs from COVID-19 convalescent donors that have been shown to decrease viral RNA in the lungs of animal models66,67. These and other mAb treatments should be entering clinical trials soon and are one of the most promising treatments against COVID-19 that have the potential to be highly efficacious and can be scaled up rapidly.
6. Correlates of protection against COVID-19
The term “correlates of protection” was originally coined in the context of vaccines to refer to “an immune responses that is responsible for and statistically interrelated with protection” against a pathogen68. Since most current vaccines induce serum antibodies, it commonly refers to levels of antibodies generated after vaccination that lead to protection from infection. For many vaccines, these levels have been determined empirically, either by challenging vaccinated individuals with the pathogen or by large scale studies following vaccination to determine the actual titer of antibody that confers protection. For example, 0.01-0.1IU/ml and 0.5IU/ml serum antibody are considered correlates of protection against diptheria69 and rabies70, respectively. For the seasonal flu vaccine an antibody titer of 1:40 in the hemagglutinin inhibition assay is considered protective71. In the case of vaccines against some pathogens such as varicella zoster and Mycobacterium tuberculosis, the correlates of protection are known to be CD4+ T cells72 and interferon levels73 respectively, but the actual amounts that confer protection has not been deduced.
Outside of vaccines, natural infections also lead to protection against subsequent exposures, therefore correlates of protection are relevant in this context as well. In the current COVID-19 pandemic, the notion of an “immunity passport” has been a popular topic of discussion, referring to the concept that by scaling up antibody testing, it may be possible to determine who is immune to COVID-19, which would provide estimates of a) herd immunity and b) who can re-enter the workforce without fear of being infected47. However, this discussion is premature as the correlates of protection against COVID-19 are currently not known, although studies examining antibody levels and functions are starting to provide more information74. While almost all individuals infected with SARS-CoV-2 develop antibodies, majority (80-90%) of them had low overall antibody levels and neutralizing antibody titers49. The antibody response also appears to be correlated with severity of the disease, which implies that in individuals who had a mild infection, the immune response may have been weak, and the antibody response may not be strong enough to confer protection29,30. Furthermore, recent studies have shown that most people who develop the disease develop neutralizing antibodies, but the nAb levels start decreasing within 8 weeks49,75.
The contribution of T cells to protection against COVID-19 re-infection is still not known and it is possible that they also contribute to protection. Another concern with coronaviruses is that many of them induce inflammatory immune responses that may exacerbate the disease and both T cells and antibodies can contribute to this12. Therefore, much more detailed studies are needed to measure immune responses from individuals across the disease spectrum. Preliminary data show that rhesus macaque monkeys who were re-challenged after recovering from primary SARS-CoV-2 infection were protected. This is promising since the protection correlated with a strong nAb response in the monkeys74. Longitudinal studies that follow individuals who have recovered from COVID-19 for subsequent re-infections need to be conducted and levels of antibodies will need to be correlated with re-infections. With these studies we will be able to determine if indeed antibodies are sufficient and if so, what levels are needed to confer protection.
7. Deleterious effects of immune responses to coronaviruses
Although immune responses to viruses play a critical role in clearing the infection and protecting from re-infection, pro-inflammatory immune responses can also be associated with disease and severe outcomes, especially following respiratory viral infections. Respiratory syncytial virus (RSV), Influenza virus, and adenovirus can all lead to pneumonia associated with host pro-inflammatory immune responses76. SARS-CoV-1 and MERS-CoV have also been associated with overexuberant immune responses that can lead to severe disease with symptoms of fever and pneumonia followed by acute respiratory distress syndrome (ARDS), thought to be due to cytokine release syndrome (CRS)77. About 20% of SARS-CoV-2 infected patients develop severe symptoms and about 5% develop acute respiratory distress14,78–81. In patients with co-morbidities such as cardiovascular disease the proportion of patients with morbidity is higher82. Like other coronavirus diseases, the severe disease in SARS-CoV-2 develops 7-10 days after symptoms develop, a timing associated with uncontrolled overexuberant host responses, rather than a direct effect of viremia83. Although the symptoms associated with SARS-CoV-2 acute respiratory distress are similar to CRS, the underlying mechanism leading to this condition is still being studied. We now know that SARS-CoV-2 infects a variety of host cells including those in the airways, cardiovascular tissue as well as immune cells, including monocytes, macrophages and possibly dendritic cells. Patients with severe disease have elevated levels of C-reactive protein (CRP), IL-6 and other inflammatory cytokines in their serum and these are associated with worse clinical outcomes84,85. Lymphopenia characterized by decreased levels of CD4, CD8 and NK cells correlated with severity of COVID-1986. T cell markers consistent with exhaustion and presence of inflammatory monocytes have also been reported18.
In addition to T cell associated inflammation, there are also concerns that antibody response to SARS-CoV-2 may also contribute to respiratory pathology due to ADE. Other coronaviruses like SARS-CoV-1 were also associated with ADE, where higher antibody titers were associated with worse morbidity and death87,88. ADE is a phenomenon associated with presence of non-neutralizing IgG antibodies that bind to the virus and then to cells that express a specific antibody receptor (Fc-receptor). This leads to activation of some cells, like monocytes and macrophages, which express Fc-receptor, production of inflammatory cytokines, and the accumulation of these cells in the lungs leading to injury89. Studies are ongoing to determine if ADE is observed in patients with severe outcomes of COVID-19.
In summary, COVID-19 patients with severe disease exhibit acute respiratory distress related to CRS, possibly caused by uncontrolled inflammation. The underlying cause maybe the activation of a number of immune cells including monocytes, macrophages, dendritic cells and T cells and the absence or inability of normal mechanisms that shut off the immune responses. Recent studies showing benefit from the use of dexamethasone, a corticosteroid which is immunosuppressive, in patients with severe disease supports the conclusion that uncontrolled inflammation underlies morbidity in COVID-1990.
8. Summary
SARS-CoV-2 is a newly emergent coronavirus that is highly infectious and is the cause of COVID-19. The disease caused by the virus is characterized primarily by mild to severe respiratory symptoms that can lead to hospitalization and death in ~1% of cases. Diagnostic tests for detection of the virus in the airways were developed quickly, within weeks of the outbreak, and have played a key role in public health strategy used globally to test, track and isolate cases, likely saving millions of lives worldwide. However, a lot more research is needed to understand this new virus, the disease, and the immune response that could potentially protect, or contribute, to COVID-19. Based on rapidly emerging data, SARS-CoV-2 infection induces differential disease, which is likely the outcome of the interaction between the virus and host immune response (Figure). The most severe disease outcomes are correlated with an over-exuberant immune response. The role of antibodies in immune protection is still not completely understood, making predictions around herd immunity uncertain and utility of “immunity passports” difficult to assess. The final exit strategy from the current pandemic will very likely involve a combination of effective therapeutics to decrease the burden of disease and vaccines to prevent transmission.
Bibliography
1. Woo, P. C. Y., Lau, S. K. P., Huang, Y. & Yuen, K.-Y. Coronavirus Diversity, Phylogeny and Interspecies Jumping. Exp. Biol. Med. 234, 1117–1127 (2009).
2. Braciale, T. J. & Hahn, Y. S. Immunity to viruses. Immunological Reviews vol. 255 5–12 (2013).
3. Duan, L. & Mukherjee, E. Focus: Microbiome: Janeway’s Immunobiology, Ninth Edition. Yale J. Biol. Med. 89, 424 (2016).
4. Cui, J., Li, F. & Shi, Z. L. Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology vol. 17 181–192 (2019).
5. Gorbalenya, A. E. et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology vol. 5 536–544 (2020).
6. Tang, X. et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. nwaa036 (2020) doi:10.1093/nsr.
7. Jiang, S., Hillyer, C. & Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends in Immunology vol. 41 355–359 (2020).
8. Lu, R. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 (2020).
9. Wan, Y., Shang, J., Graham, R., Baric, R. S. & Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 94, (2020).
10. Du, M. et al. Multiomics Evaluation of Gastrointestinal and Other Clinical Characteristics of COVID-19. Gastroenterology 158, 2298-2301.e7 (2020).
11. Grifoni, A. et al. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 27, 671-680.e2 (2020).
12. Vabret, N. et al. Immunology of COVID-19: Current State of the Science. Immunity 52, 910–941 (2020).
13. Blanco-Melo, D. et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, (2020).
14. Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
15. Hadjadj, J. et al. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. medRxiv 2020.04.19.20068015 (2020) doi:10.1101/2020.04.19.20068015.
16. Wang, F. et al. Characteristics of Peripheral Lymphocyte Subset Alteration in COVID-19 Pneumonia. J. Infect. Dis. 221, 1762–1769 (2020).
17. Zheng, M. et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cellular and Molecular Immunology vol. 17 533–535 (2020).
18. Diao, B. et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 11, 827 (2020).
19. Liu, B. et al. Persistent SARS-CoV-2 presence is companied with defects in adaptive immune system in non-severe COVID-19 patients. medRxiv http://medrxiv.org/lookup/doi/10.1101/2020.03.26.20044768 (2020) doi:10.1101/2020.03.26.20044768.
20. Thevarajan, I. et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nature Medicine vol. 26 453–455 (2020).
21. Zhou, Y. et al. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl. Sci. Rev. nwaa041 (2020).
22. Xu, Z. et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 8, 420–422 (2020).
23. Moore, J. B. & June, C. H. Cytokine release syndrome in severe COVID-19. Science. 368, 473–474 (2020).
24. Huang, A. T. et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv 2020.04.14.20065771 (2020) doi:10.1101/2020.04.14.20065771.
25. Amanat, F. et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 1–4 (2020) doi:10.1038/s41591-020-0913-5.
26. To, K. K. W. et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 20, 565–574 (2020).
27. Ju, B. et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 1–8 (2020) doi:10.1038/s41586-020-2380-z.
28. Wu, F. et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv (2020).
29. Cao, W. C., Liu, W., Zhang, P. H., Zhang, F. & Richardus, J. H. Disappearance of antibodies to SARS-associated coronavirus after recovery. New England Journal of Medicine vol. 357 1162–1163 (2007).
30. Callow, K. A., Parry, H. F., Sergeant, M. & Tyrrell, D. A. J. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 105, 435–446 (1990).
31. Wu, L. P. et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 13, 1562–1564 (2007).
32. Ko, J. H. et al. Serologic responses of 42 MERS-coronavirus-infected patients according to the disease severity. Diagn. Microbiol. Infect. Dis. 89, 106–111 (2017).
33. Payne, D. C. et al. Persistence of antibodies against middle east respiratory syndrome coronavirus. Emerg. Infect. Dis. 22, 1824–1826 (2016).
34. Li, Q. et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 382, 1199–1207 (2020).
35. Alimohamadi, Y., Taghdir, M. & Sepandi, M. Estimate of the Basic Reproduction Number for COVID-19: A Systematic Review and Meta-analysis. J. Prev. Med. Public Heal. 53, 151–157 (2020).
36. “When will it be over?”: An introduction to viral reproduction numbers, R0 and Re - CEBM. https://www.cebm.net/covid-19/when-will-it-be-over-an-introduction-to-viral-reproduction-numbers-r0-and-re/.
37. D’Arienzo, M. & Coniglio, A. Assessment of the SARS-CoV-2 basic reproduction number, R0, based on the early phase of COVID-19 outbreak in Italy. Biosaf. Heal. (2020) doi:10.1016/j.bsheal.2020.03.004.
38. Biggerstaff, M., Cauchemez, S., Reed, C., Gambhir, M. & Finelli, L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect. Dis. 14, 480 (2014).
39. Guerra, F. M. et al. The basic reproduction number (R0) of measles: a systematic review. The Lancet Infectious Diseases vol. 17 e420–e428 (2017).
40. Oran, D. P. & Topol, E. J. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann. Intern. Med. (2020) doi:10.7326/m20-3012.
41. Loeffelholz, M. J. & Tang, Y. W. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerging Microbes and Infections vol. 9 747–756 (2020).
42. Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25, (2020).
43. Dhama, K. et al. Coronavirus Disease 2019–COVID-19. Clin. Microbiol. Rev. 33, (2020).
44. Wölfel, R. et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020).
45. Serology Testing for COVID-19 at CDC | CDC. https://www.cdc.gov/coronavirus/2019-ncov/lab/serology-testing.html.
46. Xiao, S., Wu, Y. & Liu, H. Evolving status of the 2019 novel coronavirus infection: Proposal of conventional serologic assays for disease diagnosis and infection monitoring. J. Med. Virol. 92, 464–467 (2020).
47. ‘Immunity passports’ in the context of COVID-19. https://www.who.int/news-room/commentaries/detail/immunity-passports-in-the-context-of-covid-19.
48. Krammer, F. & Simon, V. Serology assays to manage COVID-19. Science. 368, 1060–1061 (2020).
49. Long, Q.-X. et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 1–5 (2020) doi:10.1038/s41591-020-0965-6.
50. Gomes, M. G. M. et al. Individual variation in susceptibility or exposure to SARS-CoV-2 lowers the herd immunity threshold. medRxiv http://medrxiv.org/lookup/doi/10.1101/2020.04.27.20081893 (2020) doi:10.1101/2020.04.27.20081893.
51. Haveri, A. et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Eurosurveillance vol. 25 (2020).
52. Thompson, C. et al. Neutralising antibodies to SARS coronavirus 2 in Scottish blood donors - a pilot study of the value of serology to determine population exposure. medRxiv http://medrxiv.org/lookup/doi/10.1101/2020.04.13.20060467 (2020) doi:10.1101/2020.04.13.20060467.
53. Immune responses and immunity to SARS-CoV-2. https://www.ecdc.europa.eu/en/covid-19/latest-evidence/immune-responses.
54. Sero-surveillance of COVID-19 - GOV.UK. https://www.gov.uk/government/publications/national-covid-19-surveillance-reports/sero-surveillance-of-covid-19.
55. Stringhini, S. et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet 0, (2020).
56. Xu, X. et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat. Med. 1–3 (2020) doi:10.1038/s41591-020-0949-6.
57. Sood, N. et al. Seroprevalence of SARS-CoV-2–Specific Antibodies Among Adults in Los Angeles County, California, on April 10-11, 2020. JAMA 323, 2425 (2020).
58. Ahn, J. Y. et al. Use of convalescent plasma therapy in two covid-19 patients with acute respiratory distress syndrome in Korea. J. Korean Med. Sci. 35, (2020).
59. Duan, K. et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 117, 9490–9496 (2020).
60. Shen, C. et al. Treatment of 5 Critically Ill Patients with COVID-19 with Convalescent Plasma. JAMA - J. Am. Med. Assoc. 323, 1582–1589 (2020).
61. Zhang, B. et al. Treatment With Convalescent Plasma for Critically Ill Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Chest (2020) doi:10.1016/j.chest.2020.03.039.
62. Li, L. et al. Effect of Convalescent Plasma Therapy on Time to Clinical Improvement in Patients With Severe and Life-threatening COVID-19. JAMA (2020) doi:10.1001/jama.2020.10044.
63. Marovich, M., Mascola, J. R. & Cohen, M. S. Monoclonal Antibodies for Prevention and Treatment of COVID-19. JAMA (2020) doi:10.1001/jama.2020.10245.
64. Marston, H. D., Paules, C. I. & Fauci, A. S. Monoclonal antibodies for emerging infectious diseases - Borrowing from history. New England Journal of Medicine vol. 378 1469–1472 (2018).
65. Mulangu, S. et al. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 381, 2293–2303 (2019).
66. Cao, Y. et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 182, 1–12 (2020).
67. Rogers, T. F. et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science (2020) doi:10.1126/science.abc7520.
68. Plotkin, S. A. Correlates of protection induced by vaccination. Clinical and Vaccine Immunology vol. 17 1055–1065 (2010).
69. Gowin, E. et al. Does vaccination ensure protection? Assessing diphtheria and tetanus antibody levels in a population of healthy children:A cross-sectional study. Med. (United States) 95, e5571 (2016).
70. Moore, S. M. et al. Rabies virus antibodies from oral vaccination as a correlate of protection against lethal infection in wildlife. Trop. Med. Infect. Dis. 2, (2017).
71. Coudeville, L. et al. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: Development and application of a bayesian random-effects model. BMC Med. Res. Methodol. 10, (2010).
72. Lal, H. et al. Efficacy of an Adjuvanted Herpes Zoster Subunit Vaccine in Older Adults. N. Engl. J. Med. 372, 2087–2096 (2015).
73. Flynn, J. A. L. et al. An essential role for interferon γ in resistance to mycobacterium tuberculosis infection. J. Exp. Med. 178, 2249–2254 (1993).
74. Yu, J. et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. eabc6284 (2020) doi:10.1126/science.abc6284.
75. Adams, E. R. et al. Evaluation of antibody testing for SARS-Cov-2 using ELISA and lateral flow immunoassays. medRxiv http://medrxiv.org/lookup/doi/10.1101/2020.04.15.20066407 (2020) doi:10.1101/2020.04.15.20066407.
76. Newton, A. H., Cardani, A. & Braciale, T. J. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Seminars in Immunopathology vol. 38 471–482 (2016).
77. Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. The Lancet vol. 395 1033–1034 (2020).
78. Guan, W. et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020).
79. Wang, D. et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA - J. Am. Med. Assoc. 323, 1061–1069 (2020).
80. Yang, X. et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 8, 475–481 (2020).
81. Murthy, S., Gomersall, C. D. & Fowler, R. A. Care for Critically Ill Patients with COVID-19. JAMA - Journal of the American Medical Association vol. 323 1499–1500 (2020).
82. Clark, A. et al. Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: a modelling study. Lancet Glob. Heal. 0, 1–15 (2020).
83. Tay, M. Z., Poh, C. M., Rénia, L., MacAry, P. A. & Ng, L. F. P. The trinity of COVID-19: immunity, inflammation and intervention. Nature Reviews Immunology vol. 20 363–374 (2020).
84. Huang, Y. et al. Clinical characteristics of 36 non-survivors with COVID-19 in Wuhan, China. medRxiv https://www.medrxiv.org/content/10.1101/2020.02.27.20029009v1 (2020) doi:10.1101/2020.02.27.20029009.
85. Xiang, J. et al. Potential biochemical markers to identify severe cases among COVID-19 patients. medRxiv vol. 19 http://medrxiv.org/lookup/doi/10.1101/2020.03.19.20034447 (2020).
86. Fei, J. et al. Reduction of lymphocyte at early stage elevates severity and death risk of COVID-19 patients: a hospital-based case-cohort study. medRxiv 2020.04.02.20050955 (2020) doi:10.1101/2020.04.02.20050955.
87. Yip, M. S. et al. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 11, 82 (2014).
88. Liu, L. et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI insight 4, (2019).
89. Taylor, A. et al. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 268, 340–364 (2015).
90. Horby, P. et al. Effect of Dexamethasone in Hospitalized Patients with COVID-19: Preliminary Report. medRxiv 2020.06.22.20137273 (2020) doi:10.1101/2020.06.22.20137273.
This article initially appeared in the Globe and Mail on July 6, 2020.



