The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019 led to the third international outbreak caused by a high consequence coronavirus in the past two decades, and the first confirmed coronavirus pandemic in recorded history. Although SARS-CoV and Middle East respiratory syndrome (MERS)-CoV strongly hinted at the potential danger of coronaviruses, few imagined the scale of the global impact of SARS-CoV-2. The question “What makes some coronaviruses such a threat to global health?” is on the minds of virologists, immunologists, epidemiologists, clinicians and many others in diverse disciplines. Here we discuss some of the remarkable aspects of coronavirus biology and what we have learned about SARS-CoV-2 since its emergence just over a year and a half ago.
Coronaviruses are a highly diverse group of viruses belonging to the family Coronaviridae. These viruses infect a wide range of species, including birds and mammals. The ‘common cold’ viruses that cause relatively mild upper respiratory tract infections (V’Kovski et al., 2021) originally circulated in animal reservoir species, and humans became infected either through contact with the reservoir species or an intermediate animal host (sometimes livestock species) (Cui et al., 2019). For example, the common cold coronavirus OC43 is thought to have originated in rodents and spilled over into humans through a bovine (cow) intermediate at some point in the distant past (Bidokhti et al., 2013). Similarly, some data suggest that certain bat species are the ancestral reservoir of MERS-CoV, and that the virus was transmitted from camels to humans. SARS-CoV-2 likely originated in bats as well, before spilling over into humans (Banerjee et al., 2021; Zhou et al., 2021). This inference is based on the close resemblance of the original SARS-CoV-2 genome to those of several related coronaviruses detected in bats (Wacharapluesadee et al., 2021; Zhou et al., 2021). At this time, details of the route of transmission remain elusive, and it is not known whether an intermediate host was involved. [SM1]
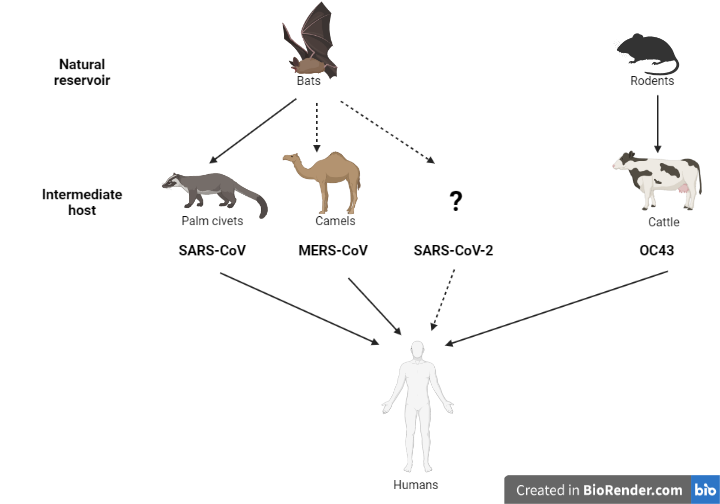
These zoonotic spillover case studies underscore the importance of improving our global understanding of viral ecology (where viruses live). Only an exceedingly small number of viruses will have the capacity to pose a risk to other species, but the implications of spillover are significant, and these rare, high impact events will likely increase in frequency in decades to come. Understanding the viral determinants of inter-species transmission, as well as the environmental factors and human behaviours that facilitate zoonotic spillovers will enhance our ability to anticipate and prevent such events from happening. Our current understanding of coronavirus biology has benefited greatly f\om the SARS-CoV-2 research, so we have a much better sense of what to look for in newly-discovered coronaviruses, and are better informed about the potential risks they may pose to human and animal health.
SARS-CoV-2 is associated with a spectrum of illness, ranging from asymptomatic or mild-moderate infection (the majority) to viral pneumonia and respiratory failure requiring hospitalization, intensive care unit admission and/or death, particularly among adults. Presenting symptoms may include fever, cough, shortness of breath as well as a range of less common manifestations such as loss of smell or taste. In addition, SARS-CoV-2 is associated with post-COVID-19 conditions, also known as long COVID, which can occur in both younger and older patients.
As SARS-CoV-2 continues to spread and adapt in the human population, it will be important to observe its epidemiological and evolutionary trajectories. Many key questions remain. Will SARS-CoV-2 disappear like SARS-CoV? Will SARS-CoV-2 cause ongoing but sporadic cases like MERS-CoV? Will SARS-CoV-2 become another endemic, seasonal respiratory virus circulating among humans and other susceptible species? Will it become more, or less pathogenic? As SARS-CoV-2 continues to defy the boundaries of our knowledge and experience, we must prepare for any and all possibilities.
- Viral building blocks[SM2]
The genomes of coronaviruses are remarkably large relative to other RNA viruses, consisting of approximately 30 kilobases (30, 000 nucleotides) (Gorbalenya et al., 2006); in contrast, the influenza virus genome is only approximately 13.5 kilobases long. The genomes of coronaviruses are positive-sense single-stranded RNA molecules, and the genetic code (RNA) is oriented such that it can be directly read by ribosomes, which are the protein factories in cells translating genes into functional proteins. The SARS-CoV-2 genome encodes sixteen non-structural proteins (nsps) primarily used for viral replication (making more viruses), and nine accessory proteins used by the virus to skirt host defenses, among other functions not yet fully understood. The virus also produces four structural proteins, the nucleocapsid (N), spike (S), envelope (E), and membrane (M) proteins. The N protein coats the RNA genome, both of which are surrounded by a protective envelope derived from host cells before being released. The S protein gives coronaviruses the distinct appearance of a crown or a ‘corona’ on the viral surface. The M protein is the most abundant one on the surface of the virion and together with the E protein, plays an important role in the formation of new virions (Neuman et al., 2011; Schoeman and Fielding, 2019).
- Unlocking the door
SARS-CoV-2 must enter a host cell and hijack the cellular machinery to copy its genetic material (replication) and generate viral proteins (translation) to eventually produce progeny virions. The S protein, acts as a ‘master key’ for a corresponding ‘lock’ or receptor and mediates entry into host cells. Different viruses use different receptors to facilitate entry, which partly determines the cells they can infect (tropism).
The S protein is composed of an upper globular structure (S1) which includes the receptor binding domain (RBD; the bit of the key) that engages with the host receptor (the lock), and a lower stalk region (S2) that is responsible for viral fusion and entry (Shang et al., 2020; Wang et al., 2020). The S protein, which is one of the more variable regions of the virus, protrudes from the surface and is under selective pressure from the host’s immune system, as it is an attractive target for antibodies to bind to and prevent entry into cells. However, certain mutations in the RBD may not only reduce how tightly neutralizing antibodies bind but may also improve the ‘fit’ of the S key, permitting more efficient cell entry. The SARS-CoV-2 RBD specifically recognizes angiotensin-converting enzyme 2 (ACE2) as its receptor on host cells (Lan et al., 2020), and once attached initiates a series of events leading to viral infection.
While the interaction between the RBD and ACE2 is critically important, it is not sufficient for virus entry, just like inserting a key into a lock is not enough to open a door. To release the viral genome into a cell and cause infection, there is a sequence of steps which involve snipping the S protein in two different places. Host proteases function like a pair of molecular scissors, and while the first snip separates S1 from S2, the second liberates the S2 fusion machinery, enabling it to fuse the cellular membrane with the viral envelope. SARS-CoV-2 S has a polybasic cleavage site not present in SARS-CoV, which allows a broader range of protease activity for the first cleavage event and may be one of the reasons SARS-CoV-2 appears to transmit more efficiently than SARS-CoV (Peacock et al., 2021). Next, the membrane envelope surrounding the virus and the host cell membrane need to fuse and form an opening through which the viral genome can pass to enter the cell. This step is also enabled by molecular scissors produced by the host cell and may involve different proteases. One important protease is called transmembrane protease, serine 2 (TMPRSS2), which snips another part of the S protein (S’), exposing the fusion domain of the S protein and activating its spring-loaded function, turning the key and bringing the viral and cellular membranes close together to ‘open the door’ by forming a channel between the two membranes, liberating the viral genome in the cell. (Hoffmann et al., 2020) (Lan et al., 2020).
- The molecular machine
Once the SARS-CoV-2 genome is inside the cell, a complex program of gene expression is initiated, whereby the virus reads and copies its genes to make transcripts (messages that are translated into viral proteins) to generate full-length genome copies to package into new viruses. This is accomplished using a viral molecular machine called an RNA-dependent RNA polymerase (RdRp). While some spontaneous mutations may allow RNA viruses to quickly adapt under selective pressure, too many errors can affect viral fitness, necessitating another viral molecular machine called an exoribonuclease (ExoN) (Denison et al., 2011) to act as a proof reader, replacing incorrect code with the correct one. Like an unwelcome guest, coronaviruses rearrange the intracellular furniture to better suit their needs, first by remodeling the cell to form circular and interconnected double membrane vesicles or pouches where virus replication and transcription occur. Our cells have internal sensors that are constantly looking for danger signals. Our own cells do not form the double stranded RNA (dsRNA) intermediate produced during virus replication and its presence would normally kickstart a cascade of antiviral responses, but this response is blunted since the viral genome within the vesicles is protected from the antiviral immune sensors of the host cell (Hagemeijer et al., 2010).
Once all of the components of the virus have been synthesized, nucleocapsids (viral RNA nestled in N proteins) are assembled into virions near the Golgi apparatus – the cellular delivery system which normally sorts and processes host proteins. (Sicari et al., 2020). The newly formed SARS-CoV-2 virions are then released from the cell and initiate infection in neighbouring cells or expelled into the environment through the nose and mouth. While attachment and entry may occur within minutes, the entire replication cycle takes several hours.
- Initial defence tactics
Coronaviruses have evolved several mechanisms to efficiently evade the innate immune responses (the entire security system) at multiple levels. For example, non-structural protein 1 (nsp1) is a key SARS-CoV-2 factor that selectively shifts gene copying machinery to suit viral needs and away from the production of antiviral proteins by host cells (Schubert et al., 2020; Thoms et al., 2020). Other accessory genes are also involved in fine tuning virus-host interactions and associated immune responses (Michel et al., 2020). Some host genes involved in antiviral responses, such as interferons (IFNs), are targets of viral proteins. IFNs modify viral pathogenicity and the severity of disease by ‘interfering ’ with viral replication and warning nearby cells to increase their defenses until proteins, known as cytokines that recruit immune cells are produced. By circumventing this response, the virus can replicate relatively unchecked. One striking feature of SARS-CoV-2 infection is its ability to block IFN responses while still activating the production of other cytokines (Blanco-Melo et al., 2020). As a result, inflammatory immune cells arrive at the site of infection, produce additional inflammatory mediators which leads to immune-mediated tissue damage (immunopathology) (Halpin et al., 2020; Matthay and Thompson, 2020). We have made great progress in understanding the initial complex interplay between the virus and the host but there is much to be learned about the full range of SARS-CoV-2 – immune system interactions. This is critically important when developing medical countermeasures such as therapeutics and vaccines.
- Mutations, mutations, mutations
Variants of concern, also known as VOCs, arise when mutations in the viral genome confer enhanced abilities to escape host immunity, transmit, or cause disease. In Canada VOCs have pushed the boundaries of the pandemic response, driving a massive third wave of viral activity. In late November 2020, a fast-spreading SARS-CoV-2 variant (B.1.1.7 or Alpha variant) with a mutation in the S protein, N501Y, was identified (Davies et al., 2021). Another variant, B.1.351 (Beta), with the same mutation in S, arose independently (Muik et al., 2021). Additional mutations such as E484K can evade virus-blocking antibodies (Andreano et al., 2020), and is seen in both Beta and Gamma (P.1) variants. The variant B.1.617.2 (Delta) appears to be more transmissible than Alpha and has been linked to the recent and devastating surge in infections in several jurisdictions as well as more severe disease (Cherian et al., 2021; Sheikh et al., 2021; Wall et al., 2021). The Delta variant contains multiple mutations in the S protein, some of which are associated with higher transmissibility, while others may allow it to evade immunity (Planas et al., 2021), underscoring the importance of ongoing genomic surveillance to detect, understand, and respond to VOCs in prescient manner.
Back to basics
Outbreaks of diseases caused by different types of viruses, such as Ebola (2013-2016), Zika (2016) and SARS-CoV-2 (ongoing), have resulted from inter-species viral spillover, and there is little doubt we will continue to witness similar events in the future. Human activities have resulted in increased contact with reservoir hosts and facilitated zoonotic disease transmission, so thoughtful consideration of how we interact with the natural world, including protecting wildlife habitats and conserving biodiversity, is key to preventing future pandemics. However, as there are approximately 1.67 million viruses thought to infect mammals and birds, it is critical that we undertake safe and comprehensive research, from the most fundamental virology perspective to population and biosphere frames of reference (Carroll et al., 2018). The molecular, ecological, epidemiological and social science data generated by these efforts will guide informed policy decisions and, together with an improved public health capacity to intervene, help prevent the emergence and transmission of novel pathogens at the human-animal interface.
Figure legends
Figure 1. Increasing our knowledge of viral ecology at the global level will not only help address important gaps in our understanding but also help us anticipate future zoonoses.
Figure 2. Key steps in the SARS-CoV-2 include viral attachment and entry into the host cell, production of viral proteins through transcription and translation, and generation of new viral genome copies through replication. Packaging and assembly is the process by which viral genomes and proteins are bundled together into virions prior to their release from the cell. (S, spike; M, membrane; N, nucleocapsid; E, envelope; RBD, receptor binding domain; TMPRSS2, transmembrane protease, serine 2).
References
Andreano, E., Piccini, G., Licastro, D., Casalino, L., Johnson, N.V., Paciello, I., Monego, S.D., Pantano, E., Manganaro, N., Manenti, A., et al. (2020). SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. Cold Spring Harbor Laboratory.
Banerjee, A., Doxey, A.C., Mossman, K., and Irving, A.T. (2021). Unraveling the Zoonotic Origin and Transmission of SARS-CoV-2. Trends in Ecology & Evolution 36, 180-184. 10.1016/j.tree.2020.12.002.
Bidokhti, M.R.M., Tråvén, M., Krishna, N.K., Munir, M., Belák, S., Alenius, S., and Cortey, M. (2013). Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains. Journal of General Virology 94, 2036-2049. 10.1099/vir.0.054940-0.
Blanco-Melo, D., Nilsson-Payant, B.E., Liu, W.-C., Uhl, S., Hoagland, D., Møller, R., Jordan, T.X., Oishi, K., Panis, M., Sachs, D., et al. (2020). Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036-1045.e1039. 10.1016/j.cell.2020.04.026.
Carroll, D., Daszak, P., Wolfe, N.D., Gao, G.F., Morel, C.M., Morzaria, S., Pablos-Méndez, A., Tomori, O., and Mazet, J.A.K. (2018). The Global Virome Project. Science 359, 872-874. 10.1126/science.aap7463.
Cherian, S., Potdar, V., Jadhav, S., Yadav, P., Gupta, N., Das, M., Rakshit, P., Singh, S., Abraham, P., and Panda, S. (2021). Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Cold Spring Harbor Laboratory.
Cui, J., Li, F., and Shi, Z.L. (2019). Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17, 181-192. 10.1038/s41579-018-0118-9.
Davies, N.G., Abbott, S., Barnard, R.C., Jarvis, C.I., Kucharski, A.J., Munday, J.D., Pearson, C.A.B., Russell, T.W., Tully, D.C., Washburne, A.D., et al. (2021). Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372, eabg3055. 10.1126/science.abg3055.
Denison, M.R., Graham, R.L., Donaldson, E.F., Eckerle, L.D., and Baric, R.S. (2011). Coronaviruses. RNA Biology 8, 270-279. 10.4161/rna.8.2.15013.
Gorbalenya, A.E., Enjuanes, L., Ziebuhr, J., and Snijder, E.J. (2006). Nidovirales: Evolving the largest RNA virus genome. Virus Research 117, 17-37. 10.1016/j.virusres.2006.01.017.
Hagemeijer, M.C., Verheije, M.H., Ulasli, M., ShaltiëL, I.A., De Vries, L.A., Reggiori, F., Rottier, P.J.M., and De Haan, C.A.M. (2010). Dynamics of Coronavirus Replication-Transcription Complexes. Journal of Virology 84, 2134-2149. 10.1128/jvi.01716-09.
Halpin, D.M.G., Singh, D., and Hadfield, R.M. (2020). Inhaled corticosteroids and COVID-19: a systematic review and clinical perspective. European Respiratory Journal 55, 2001009. 10.1183/13993003.01009-2020.
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T.S., Herrler, G., Wu, N.-H., Nitsche, A., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271-280.e278. 10.1016/j.cell.2020.02.052.
Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., and Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215-220. 10.1038/s41586-020-2180-5.
Matthay, M.A., and Thompson, B.T. (2020). Dexamethasone in hospitalised patients with COVID-19: addressing uncertainties. The Lancet Respiratory Medicine 8, 1170-1172. 10.1016/s2213-2600(20)30503-8.
Michel, C.J., Mayer, C., Poch, O., and Thompson, J.D. (2020). Characterization of accessory genes in coronavirus genomes. Virology Journal 17. 10.1186/s12985-020-01402-1.
Muik, A., Wallisch, A.-K., Sänger, B., Swanson, K.A., Mühl, J., Chen, W., Cai, H., Maurus, D., Sarkar, R., Türeci, Ö., et al. (2021). Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science 371, 1152-1153. 10.1126/science.abg6105.
Neuman, B.W., Kiss, G., Kunding, A.H., Bhella, D., Baksh, M.F., Connelly, S., Droese, B., Klaus, J.P., Makino, S., Sawicki, S.G., et al. (2011). A structural analysis of M protein in coronavirus assembly and morphology. Journal of Structural Biology 174, 11-22. 10.1016/j.jsb.2010.11.021.
Peacock, T.P., Goldhill, D.H., Zhou, J., Baillon, L., Frise, R., Swann, O.C., Kugathasan, R., Penn, R., Brown, J.C., Sanchez-David, R.Y., et al. (2021). The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 6, 899-909. 10.1038/s41564-021-00908-w.
Planas, D., Veyer, D., Baidaliuk, A., Staropoli, I., Guivel-Benhassine, F., Rajah, M.M., Planchais, C., Porrot, F., Robillard, N., Puech, J., et al. (2021). Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 10.1038/s41586-021-03777-9.
Schoeman, D., and Fielding, B.C. (2019). Coronavirus envelope protein: current knowledge. Virology Journal 16. 10.1186/s12985-019-1182-0.
Schubert, K., Karousis, E.D., Jomaa, A., Scaiola, A., Echeverria, B., Gurzeler, L.-A., Leibundgut, M., Thiel, V., Mühlemann, O., and Ban, N. (2020). SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nature Structural & Molecular Biology 27, 959-966. 10.1038/s41594-020-0511-8.
Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., and Li, F. (2020). Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences 117, 11727-11734. 10.1073/pnas.2003138117.
Sheikh, A., McMenamin, J., Taylor, B., Robertson, C., Public Health, S., and the, E.I.I.C. (2021). SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet 397, 2461-2462. 10.1016/S0140-6736(21)01358-1.
Sicari, D., Chatziioannou, A., Koutsandreas, T., Sitia, R., and Chevet, E. (2020). Role of the early secretory pathway in SARS-CoV-2 infection. Journal of Cell Biology 219. 10.1083/jcb.202006005.
Thoms, M., Buschauer, R., Ameismeier, M., Koepke, L., Denk, T., Hirschenberger, M., Kratzat, H., Hayn, M., Mackens-Kiani, T., Cheng, J., et al. (2020). Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369, 1249-1255. 10.1126/science.abc8665.
V’Kovski, P., Kratzel, A., Steiner, S., Stalder, H., and Thiel, V. (2021). Coronavirus biology and replication: implications for SARS-CoV-2. Nature Reviews Microbiology 19, 155-170. 10.1038/s41579-020-00468-6.
Wacharapluesadee, S., Tan, C.W., Maneeorn, P., Duengkae, P., Zhu, F., Joyjinda, Y., Kaewpom, T., Chia, W.N., Ampoot, W., Lim, B.L., et al. (2021). Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nature Communications 12. 10.1038/s41467-021-21240-1.
Wall, E.C., Wu, M., Harvey, R., Kelly, G., Warchal, S., Sawyer, C., Daniels, R., Hobson, P., Hatipoglu, E., Ngai, Y., et al. (2021). Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. The Lancet 397, 2331-2333. 10.1016/s0140-6736(21)01290-3.
Wang, M.-Y., Zhao, R., Gao, L.-J., Gao, X.-F., Wang, D.-P., and Cao, J.-M. (2020). SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Frontiers in Cellular and Infection Microbiology 10. 10.3389/fcimb.2020.587269.
Zhou, H., Ji, J., Chen, X., Bi, Y., Li, J., Wang, Q., Hu, T., Song, H., Zhao, R., Chen, Y., et al. (2021). Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 10.1016/j.cell.2021.06.008.